create_miRNA_logo.pl - Creates a miRNA logo using the command line version of WebLogo 3.0
# Minimal argument call specifying all required parameters.
create_miRNA_logo.pl --input Mapping_day0_1_i_Eland_against_mature_unambiguous.txt
--id "hsa-mir-219-1 MI0000296 Homo sapiens miR-219-2 stem-loop"
--output logo.pdf
# Maximum argument call specifying all possible parameters; Several different input files may be specified
# Note that also the file containing the ambiguous hits may be specified
create_miRNA_logo.pl --output logo.png --min_length 15 --max_length 32 --max_mm 2 --strand RF
--id "hsa-mir-219-1 MI0000296 Homo sapiens miR-219-2 stem-loop"
--max_ambiguity 2 --format png --start 10 --end 50 --tempdir "/tmp"
--scale-width no --stacks-per-line 100
--input Mapping_day0_1_i_Eland_against_mature_unambiguous.txt
--input Mapping_day0_1_i_Eland_against_mature_ambiguous.txt
The input files; Several files may be specified, e.g.: --input file1 --input file2.
The input files have to be output files of the script run_Mapping or run_Multimapper.
Note that unambiguously and ambiguously mapped reads may be provided for this script. Mandatory parameter
The output file. Mandatory parameter
The id of the miRNA (reference sequence) for which the profile should be created. Mandatory parameter
Only reads mapping to the specified strand will be used. Possible values: R (reverse strand), F (forward strand), RF (both strands); default=RF
The minimum length of reads. Shorter reads will not be used. default=15
The maximum length of reads. Longer reads will not be used. default=100
The maximum number of mismatches. Reads having more mismatches will not be used. default=2
The maximum ambiguity of the hits. Hits having a higher ambiguity will be ignored. The ambiguity is an integer value which relates how often a read could be mapped with an equal good score (number of mismatches) to the reference sequence. Examples:
A read which could be mapped to the H. sapiens genome only once having two mismatches, will have a ambiguity of "1".
A read which could be mapped to the H. sapiens genome three times, always having one mismatch, will have a ambiguity of "3".
A read which could be mapped to the H. sapiens genome three times having one mismatch and one time having zero mismatches, will have a ambiguity of "1".
A read which could be mapped to the H. sapiens genome three times having one mismatch and two times having zero mismatches, will have a ambiguity of "2".
default=5
Create the logo only for a subregion of the reference sequence. Start position of the subregion. default=undef
Create the logo only for a subregion of the reference sequence. End position of the subregion. default=undef
This is a weblogo parameter. Scale the visible stack width by the fraction of symbols in the column? (i.e. columns with many gaps of unknowns are narrow.) possible values: yes/no; (default: no)
This is a weblogo parameter. Maximum number of logo stacks per logo line (default: 40)
This is a weblogo parameter. Format of output: eps (default), png, png_print, pdf, jpeg, txt; default=pdf
The path to the temporary directory. default=/tmp
Display the help pages.
The script creates a miRNA-logo using the tool weblogo 3.0. This Logos may, for example, be usefull to visualise RNA editing.
Mapping results of the script run_Mapping.pl or run_Multimapper.pl.
Note that unambiguous and ambiguous mapping results may be provided.
For example:
24688||Count=3 TACCCTGTAGATCCGAATTTGT hsa-miR-10a MIMAT0000253 Homo sapiens miR-10a 1 0 F 1 128318||Count=2 TACCCTGTAGATCCGAATTTGTG hsa-miR-10a MIMAT0000253 Homo sapiens miR-10a 1 0 F 1 150952||Count=1 TACCCTGTAGATCCTAATTTGTGT hsa-miR-10a MIMAT0000253 Homo sapiens miR-10a 1 2 R 1 212857||Count=1 TACCCTGTAGATCCAAATTTGT hsa-miR-10a MIMAT0000253 Homo sapiens miR-10a 1 1 F 1 317801||Count=1 TACCTTGTAGATCCGAATTTGTG hsa-miR-10a MIMAT0000253 Homo sapiens miR-10a 1 1 F 1 389805||Count=1 TACCCTGTATATCCGAATTTGTGG hsa-miR-10a MIMAT0000253 Homo sapiens miR-10a 1 2 F 1
Ambiguity is an important concept in the MIRO-pipeline, it is therefore crucial that this concept is properly understood. In a nutshell, ambigutiy is the number of equal good mapping positions for a single Solexa-read. Equal good in this context refers to the number of mismatches. In the MIRO-pipeline all unambiguously mapped reads have a ambiguity of "1" and they are provided in a separate output-file. All ambiguously mapped reads, on the other hand, have a ambiguity of ">=2"
Examples:
A read which could be mapped to the H. sapiens genome only once having two mismatches, will have a ambiguity of "1".
A read which could be mapped to the H. sapiens genome three times, always having only one mismatch, will have a ambiguity of "3".
A read which could be mapped to the H. sapiens genome three times having one mismatch and one time having zero mismatches, will have a ambiguity of "1".
A read which could be mapped to the H. sapiens genome three times having one mismatch and two times having zero mismatches, will have a ambiguity of "2".
The output will be a logo in the specified --format.
For example:
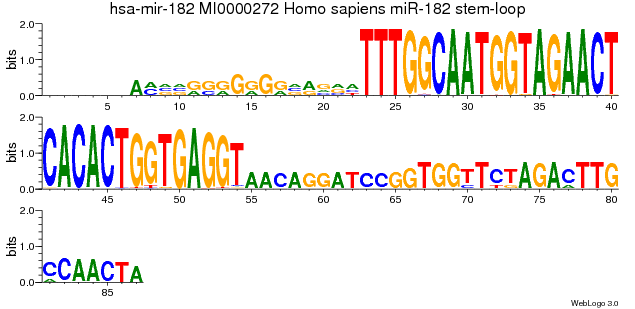
Here the parameter --scale-width has been set to no.
When using --scale-width yes the following logo would result:
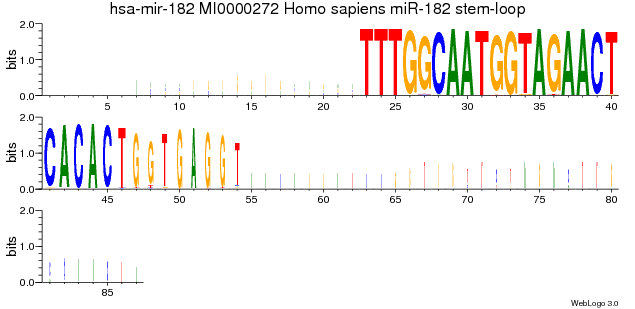
Scale-width yes reflects the actual coverage of the bases. However, the problem is that usually the coverage of the hairpin and the miRNA* is
very low when compared to the mature miRNA, this makes it very hard to identify interesting patterns in the logo.
Thus it may be useful to use --scale-width no instead, however, in the resulting graph the size of the bases does not reflect the actual coverage properly
Perl 5.8 or higher
Weblogo 3.0
Robert Kofler
Debayan Datta
Heinz Himmelbauer
robert.kofler at crg.es